Updated Cycling History map
Some (many!) years ago I posted a few images showing my riding history in California (in 2008) and Colorado (in 2012). While they are interesting, these are static images and cannot be explored.
Thirteen years have passed and technology has improved. Today I added a new page showing my entire cycling history since I started using GPS over twenty years ago. Instead of separate static images, it uses dynamic web technology™️ on a single map. As before, color shows how frequently I ride past a point on the map. However, I changed the logic: my previous maps used counts of observations near points, my new map uses counts of distinct rides that pass each point on the map. Instead of where I've done the most laps or my GPS has recorded the most times (for whatever reason), this is more indicative of where I ride the most.
For the most immersive experience, here is the full screen version.
more ...Go Big To Go Home
The first step of data science when working with new a dataset is to understand the high-level facts and relationships within the data. This is often done by exploring the data interactively by using something like Python, R, or Matlab.
Recently I've been exploring a new dataset. It's a pretty big dataset: a few hundred gigabytes of data in compressed Parquet format. A rule of thumb is that reading data off disk into memory takes ten or twenty times the memory than the storage the data uses on disk. For this dataset, that could equal more than ten terabytes of memory, which in 2025 is still a pretty ridiculous amount of memory on a single machine. It is for this reason that working with data this size requires tools that allow you to work with the data without loading all of it into memory at once.
One of these tools is Polars. Quoting the Polars home page: "Want to process large data sets that are bigger than your memory? Our streaming API allows you to process your results efficiently, eliminating the need to keep all data in memory." It's still a bit rough around the edges with some unfinished and missing features, but overall it's a powerful and capable tool for data analysis. Lately I've been using Polars more and more, taking advantage of this "streaming" ability.
On the other hand, sometimes it's easiest to do things directly and skip all the low-memory "streaming" tricks. If I can get an answer more quickly by simply using lots of memory, especially if it's something I'm doing only once and not putting into a repeated process, then this can be the right choice. Polars can do "streaming" analysis, but at a certain point it has to coalesce things into an answer, and sometimes that answer can use a significant amount of memory.
There are many negative aspects of cloud computing that I won't get into here. However, there are some good things, and one of them is that you can scale up and down resources as needed. All cloud providers, like Amazon Web Services and Google Cloud Platform offer many services and in particular Virtual Machines. When running a virtual machine you can choose the hardware specifications in terms of CPU kind and core count, amount of RAM, and other features like network speeds, GPUs, or SSDs. A virtual machine can be booted on one hardware configuration, shut down, and the rebooted on a different configuration as needed. It's as if you took the hard drive out of your laptop and put it in a big workstation. All your data and settings are still there, but you've upgraded the hardware. This is something I take advantage of frequently!
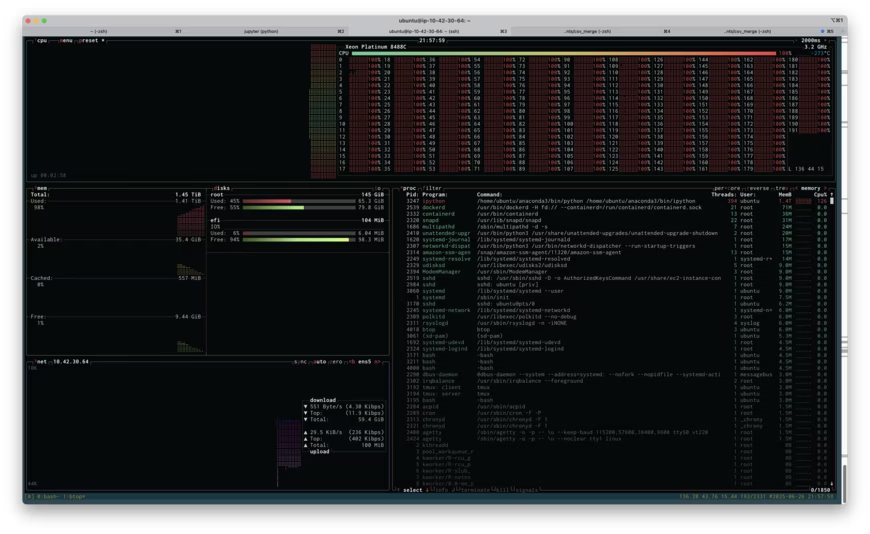
I was attempting to do a certain analysis of the new dataset on an EC2 virtual machine and I kept running out of memory. Instead of switching to some low-memory tricks, I decided to see if I could save some time by simply rebooting my virtual machine on one of the larger instances AWS offers: a r7i.48xlarge. This has 192 CPU cores and 1,536 GB of RAM. It costs $12.70 per hour. I get paid more than $12.70 per hour, so if booting up a huge machine like this saves me even a little time, it's worth it.
Above is a screenshot of btop running on the r7i.48xlarge instance while I attempted to run my analysis. If you click on the image, you'll see the full size screenshot. You'll see that I'm about to run out of memory: 1.41TB used of 1.45TB. You'll also see that I'm using all 192 cores at 100% load (the cores are labeled 0-191). Unfortunately, throwing all this memory at the problem didn't work, I ran out of memory, and I had to resort to being more clever. Being clever took more time, of course, but if the high-RAM instance had worked, it would have paid off.
Playing with various server configuration tools (like this one) shows that the r7i.48xlarge would cost at least $60,000. This is not something that I need very often, and purchasing something this large would be ridiculous. However, renting it for half an hour, if it saves me a few hours, is definitely worth it. Also, it's kind of fun to say "yeah, I used 1.5TB of memory and 192 cores and it wasn't enough."
more ...Polars scan_csv and sink_parquet
The documentation for polars is not the best,
and figuring out how to do this below took me over an hour.
Here's how to read in a headerless csv file into a
LazyFrame using
scan_csv
and write it to a parquet file using
sink_parquet.
The key is to use with_column_names and
schema_overrides.
Despite what the documentation says, using schema doesn't work as you
might imagine and sink_parquet returns with a cryptic error about
the dataframe missing column a.
This is just a simplified version of what I actually am trying to do, but that's the best way to drill down to the issue. Maybe the search engines will find this and save someone else an hour of frustration.
import numpy as np
import polars as pl
df = pl.DataFrame(
{"a": [str(i) for i in np.arange(10)], "b": np.random.random(10)},
)
df.write_csv("/tmp/stuff.csv", include_header=False)
lf = pl.scan_csv(
"/tmp/stuff.csv",
has_header=False,
schema_overrides={
"a": pl.String,
"b": pl.Float64,
},
with_column_names=lambda _: ["a", "b"],
)
lf.sink_parquet("/tmp/stuff.parquet")
Claude Code
A few weeks ago I started playing with Claude Code, which is an AI tool that can help build software projects for/with you. Like ChatGPT, you interact with the AI conversationally, using whole sentences. You can tell it what programming language and which software packages to use, and what you want the new program to do.
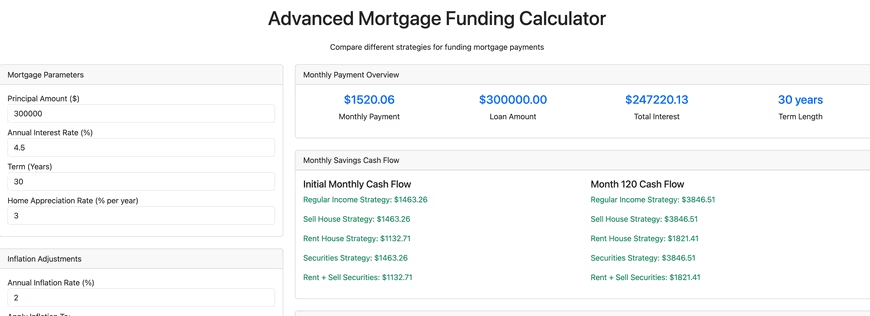
I started by asking it to build a mortgage calculator using Python and Dash. Python is one of the most popular programming languages and Dash is an open source Python package that combines Flask, a tool to build websites using Python, and Plotly, a tool that builds high-quality interactive web plots. The killer feature of Dash is that it handles all the nasty and tedious parts of an interactive webpage (meaning Javascript, eeeek!). It deals with webpage button clicks and form submissions for you, and you, the coder, can write things in lovely Python.
With a fair amount of back and forth Claude Code built this advanced mortgage calculator, which is only sorta kinda functional. It does a fair amount of what I asked it to do, but it also doesn't do a fair amount of what I asked it to do, and it has a decent number of bugs. The good things it did was handle some of the tedious boiler plate stuff like creating the necessary directory hierarchy and files, including a README.md detailing how to run the software. Creating a Dash webpage requires writing Python function(s) and a template detailing how to insert the output of the function(s) into a webpage, and Claude Code handled that with aplomb.
What it didn't handle well was more complicated things, like asking it to write an optimizer for funding/paying off a mortgage considering various funding sources and economic factors. It also didn't write the code using standard Python practices. The first time I looked at the code using VS Code and Ruff, Ruff reported over 1,000 style violations. If I found a bug in the code, some of the time I could tell Claude to fix it, and it would, but other times, it would simply fail. Altogether there's about 6,000 lines of code, and as the project got bigger it was clear that Claude was struggling. Simply put, there's a limit of the size or complexity of a codebase that Claude can handle. Humans are not going to be replaced, yet.
At this point I'm not sure what I'll do with the financial calculator. I don't think Claude can help me any more, so I'd have to manually dive in to fix bugs and improve it, and I haven't decided if I will. In summary, my impression of Claude is that it's decent at creating the base of a project or application, but then anything truly creative and complicated is beyond what it can do.
Claude Code isn't free (they give you $5 to start) and I had to deposit money to make this tool. I still have a bit of money to spend, so let's try a few tasks and see how Claude does. I've uploaded all of the code generated below to this repository.
Simple Calculator
I asked it to create a simple web-based calculator, and at a cost of $0.17, here is what it created that works well (that's not a picture below, try clicking on the buttons!).
Simple C Hello World!
Create a template for a program in C. Include a makefile and a main.c file with a main function that prints a simple "hello world!". $0.08 later it performed this simple task flawlessly.
Excel Telescope Angular Resolution
Create an Excel file that can calculate the angular resolution of a reflector telescope as a function of A) the diameter of the primary mirror and B) the altitude of the telescope. It went off and thought for a bit, and spent another $0.17 of my money. It output a file with a ".xlsx" extension, but the file can't be opened. Looking at the output of Claude, I suspect that this may be a file formatting issue because Claude is designed to handle text file output rather than binary.
Text to Speech
Seeing that Claude struggled with creating binary output, next I asked it to create a Jupyter notebook (Jupyter notebooks have the extension .ipynb but they're actually text files) that uses Python and any required packages and can take a block of text and use text to speech to output the text to a sound file. This succeeded ($0.12), and in particular used gTTS (Google Text To Speech) to do the heavy lifting.
Rust Parallel Pi Calculator
Write a program in Rust that uses Monte Carlo techniques to calculate pi. Use multithreading. The input to the program should be the desired number of digits of pi, and the output is pi to that many digits. $0.11 later, I got a Rust program that crashes with a "attempt to multiply with overflow" error. Not great! I could interact with Claude and ask it to try to fix the error, but I haven't.
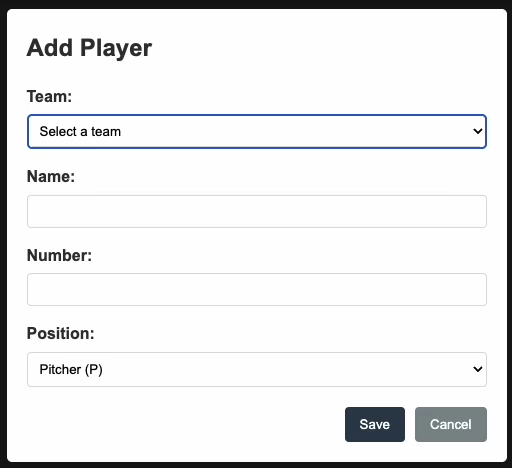
Baseball Scoresheet Web App
Create a web app for keeping score of a baseball game. Make the page resemble a baseball scoresheet. Use a SQLite database file to store the data. Make the page responsive such that each scoring action is saved immediately to the database. At a cost of $0.93, it output almost 2,000 lines of code. The resulting npm web app doesn't work. Upon initial page load it asks the user to enter player names, numbers, and positions, but no matter what you do, you cannot get past that. Bummer!
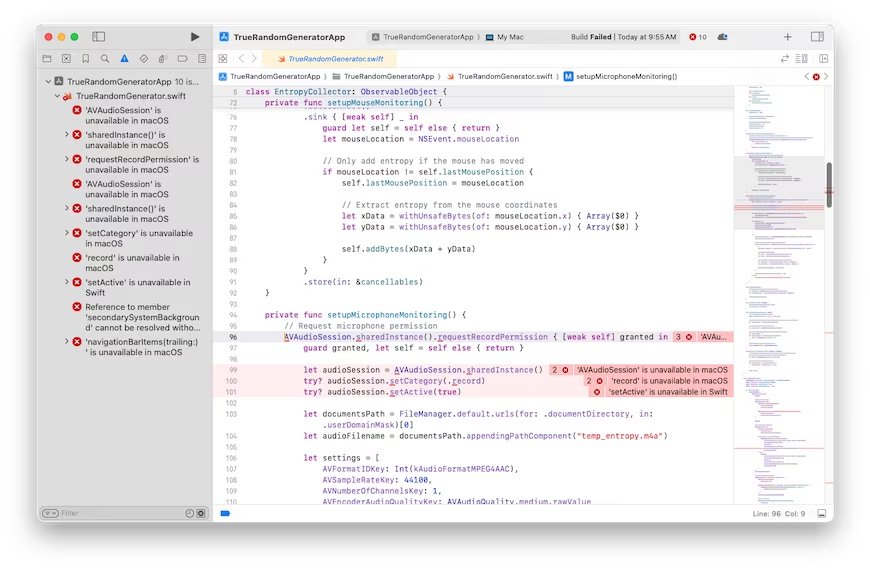
Random Number Generator
It looks like the more complicated the ask gets, the worse Claude gets. Here's one more thing I'll try that I have no idea how to do myself. We'll see how well Claude does at it. Create a Mac OS program that generates a truly random floating point number. It should not use a pseudo-random number generator. It should capture random input from the user as the source of randomness. It should give the user the option of typing random keyboard keys, or mouse movements, or making noises captured by the microphone. Please use the swift programming language. Create a Xcode compatible development stack. Create a stylish GUI that looks like a first-class Mac OS program. $0.81 later, and asking it to fix one bug, led to a second try that was also broken with 10 bugs. Clearly, I've pushed Claude past its breaking point.
I'm guessing that all of the broken code can be fixed, maybe by Claude itself, but it ultimately might require human intervention in some cases. I'm optimistic that it could fix the Rust/pi bug, but I'm not optimistic that it can fix the baseball nor random number generator stuff. AI code generation might be coming for us eventually, but not today.
more ...Setting Hadoop Node Types on AWS EMR
Amazon Web Services (AWS) offers dozens (if not over one hundred) different services. I probably use about a dozen of them regularly, including Elastic Map Reduce (EMR) which is their platform for running big-data things like Hadoop and Spark.
EMR runs your work on EC2 instances, and you can pick which kind(s) you want when the job starts. You can also pick the "lifecycle" of these instances. This means you can pick some instances to run as "on-demand" where the instance is yours (barring a hardware failure), and other instances to run as "spot" which costs much less than on-demand but AWS can take away the instance at any time.
Luckily, Hadoop and Spark are designed to work on unreliable hardware, and if previously done work is unavailable (e.g. because the instance that did it is no longer running), Hadoop and Spark can re-run that work again. This means that you can use a mix of on-demand and spot instances that, as long as AWS doesn't take away too many spot instances, will run the job for lower cost than otherwise.
A big issue with running Hadoop on spot instances is that multi-stage Hadoop jobs save some data between stages that can't be redone. This data is stored in HDFS, which is where Hadoop stores (semi)permanent data. Because we don't want this data going away, we need to run HDFS on on-demand instances, and not run it on spot instances. Hadoop handles this by having two kind of "worker" instances: "CORE" instances that run HDFS and have the important data, and "TASK" types that do not run HDFS and store easily reproduced data. Both types share in the computational workload, the difference is what kind of data is allowed to be stored on them. It makes sense, then, to confine "TASK" instances to spot nodes.
The trick is to configure Hadoop such that the instances themselves know what kind of instance they are. Figuring this out was harder than it should have been because AWS EMR doesn't auto-configure nodes to work this way; the user needs to configure the job to do this including running scripts on the instances themselves.
I like to run my Hadoop jobs using mrjob which makes development and running Hadoop with Python easier. I assume that this can be done outside of mrjob, but its up to the reader to figure out how to do that.
There are three parts to this. The first two are two Python scripts that are run on the EC2 instances, and the third is modifying the mrjob configuration file. The Python scripts should be uploaded to a S3 bucket because they will be downloaded to each Hadoop node (see the bootstrap actions below).
With the changes below, you should be able to run a multi-step Hadoop job on AWS EMR using spot nodes and not lose any intermediate work. Good luck!
make_node_labels.py
This script tells yarn what kind of instance types are available. This only needs to run once.
#!/usr/bin/python3
import subprocess
import time
def run(cmd):
proc = subprocess.Popen(cmd,
stdout = subprocess.PIPE,
stderr = subprocess.PIPE,
)
stdout, stderr = proc.communicate()
return proc.returncode, stdout, stderr
if __name__ == '__main__':
# Wait for the yarn stuff to be installed
code, out, err = run(['which', 'yarn'])
while code == 1:
time.sleep(5)
code, out, err = run(['which', 'yarn'])
# Now we wait for things to be configured
time.sleep(60)
# Now set the node label types
code, out, err = run(["yarn",
"rmadmin",
"-addToClusterNodeLabels",
'"CORE(exclusive=false),TASK(exclusive=false)"'])
get_node_label.py
This script tells Hadoop what kind of instance this is. It is called by Hadoop and run as many times as needed.
#!/usr/bin/python3
import json
k='/mnt/var/lib/info/extraInstanceData.json'
with open(k) as f:
response = json.load(f)
print("NODE_PARTITION:", response['instanceRole'].upper())
mrjob.conf
This is not a complete mrjob configuration file. It shows the essential parts needed for setting up CORE/TASK nodes. You will need to fill in the rest for your specific situation.
runners:
emr:
instance_fleets:
- InstanceFleetType: MASTER
TargetOnDemandCapacity: 1
InstanceTypeConfigs:
- InstanceType: (smallish instance type)
WeightedCapacity: 1
- InstanceFleetType: CORE
# Some nodes are launched on-demand which prevents the whole job from
# dying if spot nodes are yanked
TargetOnDemandCapacity: NNN (count of on-demand cores)
InstanceTypeConfigs:
- InstanceType: (bigger instance type)
BidPriceAsPercentageOfOnDemandPrice: 100
WeightedCapacity: (core count per instance)
- InstanceFleetType: TASK
# TASK means no HDFS is stored so loss of a node won't lose data
# that can't be recovered relatively easily
TargetOnDemandCapacity: 0
TargetSpotCapacity: MMM (count of spot cores)
LaunchSpecifications:
SpotSpecification:
TimeoutDurationMinutes: 60
TimeoutAction: SWITCH_TO_ON_DEMAND
InstanceTypeConfigs:
- InstanceType: (bigger instance type)
BidPriceAsPercentageOfOnDemandPrice: 100
WeightedCapacity: (core count per instance)
- InstanceType: (alternative instance type)
BidPriceAsPercentageOfOnDemandPrice: 100
WeightedCapacity: (core count per instance)
bootstrap:
# Download the Python scripts to the instance
- /usr/bin/aws s3 cp s3://bucket/get_node_label.py /home/hadoop/
- chmod a+x /home/hadoop/get_node_label.py
- /usr/bin/aws s3 cp s3://bucket/make_node_labels.py /home/hadoop/
- chmod a+x /home/hadoop/make_node_labels.py
# nohup runs this until it quits on its own
- nohup /home/hadoop/make_node_labels.py &
emr_configurations:
- Classification: yarn-site
Properties:
yarn.node-labels.enabled: true
yarn.node-labels.am.default-node-label-expression: CORE
yarn.node-labels.configuration-type: distributed
yarn.nodemanager.node-labels.provider: script
yarn.nodemanager.node-labels.provider.script.path: /home/hadoop/get_node_label.py
Resuscitating the Blog
When I started this blog I used Wordpress. Wordpress is web-based content management system that uses PHP and one of MySQL or MariaDB to store the post data. I used Wordpress for years moving between a few different hosts finally settling on Linode.
Wordpress is a great product and has a healthy, vibrant user community. There are thousands of plugins and themes. There's phone apps for making blog posts on-the-go. However, being a dynamic web tool it is vulnerable to security issues. It is a constant game of whack-a-mole keeping things updated. Plugins add new bugs, and sometimes plugins are written with purposeful backdoors.
Over three years ago I decided to move away from Wordpress to something non-dynamic. Something with fewer vulnerabilities. I initially tried Hugo and found a tool that exports posts from Wordpress to Hugo. However, I found I didn't really like Hugo, and I just kinda... didn't do anything about it for a long time. Years a long time.
A few weeks ago I decided to revisit things. I found Pelican, a Python-based static website builder.
It is very similar to Hugo in form and function, but being written in Python it jibes with my knowledge
and skills much better than Hugo.
Posts and pages are written in reStructuredText or Markdown, or even
HTML. I mostly use Markdown,
and in a few cases, HTML is the correct choice (e.g. photo galleries).
Each document has fields of metadata that Pelican parses to build out a full static website.
There are a number of themes and I'm presently using the bootcamp2 theme.
Pelican also has dozens of plugins that add
plenty of capabilities but zero vulnerabilities.
It took a few weeks to slowly fix up all the posts that previously had been exported from Wordpress into Hugo-flavored Markdown into something Pelican could read. I uploaded a fair number of videos to YouTube, and had to adjust various other media here and there.
At the same time I'm moving my website to a new Linode server. This new server will not run any PHP or other dynamic web content which should keep things much simpler.
I'm going to try to post here more often and more consistently. Let's see if I succeed!
more ...How I Got a 327x Speedup Of Some Python Code
I don't usually post code stuff on this blog, but I had some fun working on this and I wanted to share!
My colleague, Abhi, is processing data from an instrument collecting actual data from the real world (you should know that this is something I have never done!) and is having some problems with how long his analysis is taking. In particular, it is taking him longer than a day to analyze a day's worth of data, which is clearly an unacceptable rate of progress. One step in his analysis is to take the data from all ~30,000 frequency channels of the instrument, and calculate an average across a moving window 1,000 entries long. For example, if I had the list:
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
and I wanted to find the average over a window of length three, I'd get:
[1, 2, 3, 4, 5, 6, 7, 8]
Notice that I started by averaging [0, 1, 2]=>1, as this is the first way to come up with a window of three values. Similarly, the last entry is [7, 8, 9]=>8. Here is a simplified version of how Ahbi was doing it (all of the code snippets shown here are also available [here][1]):
def avg0(arr, l):
# arr - the list of values
# l - the length of the window to average over
new = []
for i in range(arr.size - l + 1):
s = 0
for j in range(i, l + i):
s += j
new.append(s / l)
return new
This function is correct, but when we time this simple function (using an IPython Magic) we get:
# a is [0, 1, 2, ..., 29997, 29998, 29999]
a = np.arange(30000)
%timeit avg0(a, 1000)
1 loops, best of 3: 3.19 s per loop
Which isn't so horrible in the absolute sense -- 3 seconds isn't that long compared to how much time we all waste per day. However, I neglected to mention above that Abhi must do this averaging for nearly 9,000 chunks of data per collecting day. This means that this averaging step alone takes about 8 hours of processing time per day of collected data. This is clearly taking too long!
The next thing I tried is to take advantage of Numpy, which is a Python package that enables much faster numerical analysis than with vanilla Python. In this function I'm essentially doing the same thing as above, but using Numpy methods, including pre-allocating space, summing values using the accelerated Numpy .sum() function, and using the xrange() iterator, which is somewhat faster than plain range():
def avg1(arr, l):
new = np.empty(arr.size - l + 1, dtype=arr.dtype)
for i in xrange(arr.size - l + 1):
new[i] = arr[i:l+i].sum() / l
return new
This provides a healthy speed up of about a factor of eight:
%timeit avg1(a, 1000)
1 loops, best of 3: 405 ms per loop
But this still isn't fast enough; we've gone from 8 hours to just under an hour. We'd like to run this analysis in at most a few minutes. Below is an improved method that takes advantage of a queue, which is a programming construct that allows you to efficiently keep track of values you've seen before.
def avg2(arr, l):
new = np.empty(arr.size - l + 1, dtype=arr.dtype)
d = deque()
s = 0
i = 0
for value in arr:
d.append(value)
s += value
if len(d) == l:
new[i] = s / l
i += 1
elif len(d) > l:
s -= d.popleft()
new[i] = s / l
i += 1
return new
And here we can see that we've cut our time down by another factor of 4:
%timeit avg2(a, 1000)
10 loops, best of 3: 114 ms per loop
This means that from 8 hours we're now down to roughly 13 minutes. Getting better, but still not great. What else can we try? I've been looking for an excuse to try out Numba, which is a tool that is supposed to help speed up numerical analysis in Python, so I decided to give it a shot. What makes Numba attractive is that with a single additional line, Numba can take a function and dramatically speed it up by seamlessly converting it into C and compiling it when needed. So let's try this on the first averaging function:
@jit(argtypes=[int32[:], int32], restype=int32[:])
def avg0_numba(arr, l):
new = []
for i in range(arr.size - l + 1):
s = 0
for j in range(i, l + i):
s += j
new.append(s / l)
return np.array(new)
In the line beginning with @jit, all I have to do is describe the input and the output types, and it handles the rest. And here's the result:
%timeit avg0_numba(a, 1000)
10 loops, best of 3: 21.6 ms per loop
What is incredible here is that not only is this roughly 5 times faster than the queue method above, it's a ridiculous 147 times faster than the original method and only one line has been added. We've now reduced 8 hours to about 4 minutes. Not bad!
Let's try this on the second averaging method, which if you recall, was substantially better than the original method:
@jit(argtypes=[int32[:], int32], restype=int32[:])
def avg1_numba(arr, l):
new = np.empty(arr.size - l + 1, dtype=arr.dtype)
for i in xrange(arr.size - l + 1):
new[i] = arr[i:l+i].sum() / l
return new
%timeit avg1_numba(a, 1000)
1 loops, best of 3: 688 ms per loop
That's interesting! For some reason I don't understand, this is actually slower than the un-optimized version of avg1. Let's see if Numba can speed up the queue method:
@jit(argtypes=[int32[:], int32], restype=int32[:])
def avg2_numba(arr, l):
new = np.empty(arr.size - l + 1, dtype=arr.dtype)
d = deque()
s = 0
i = 0
for value in arr:
d.append(value)
s += value
if len(d) == l:
new[i] = s / l
i += 1
elif len(d) > l:
s -= d.popleft()
new[i] = s / l
i += 1
return new
%timeit avg2_numba(a, 1000)
10 loops, best of 3: 77.5 ms per loop
This is somewhat better than before, but still not as fast as avg0_numba, which comes in at roughly 20ms. But what if I really try hard to optimize the queue method by using only Numpy arrays?
@jit(argtypes=[int32[:], int32], restype=int32[:])
def avg2_numba2(arr, l):
new = np.empty(arr.size - l + 1, dtype=arr.dtype)
d = np.empty(l + 1, dtype=arr.dtype)
s = 0
i = 0
left = 0
right = 0
full = False
for j in xrange(arr.size):
d[right] = arr[j]
s += arr[j]
right = (right + 1) % (l+1)
if not full and right == l:
new[i] = s / l
i += 1
full = True
elif full:
s -= d[left]
left = (left + 1) % (l+1)
new[i] = s / l
i += 1
return new
%timeit avg2_numba2(a, 1000)
100 loops, best of 3: 9.77 ms per loop
A ha! That's even faster, and our 3.19s are now down to 9.77ms, an improvement of 327 times. The original 8 hours are now reduced to less than two minutes. I hope you've enjoyed this as much as I have!
Update:
After chatting with a couple of my colleagues, this appears to be the fastest way to do this kind of operation:
from scipy.signal import fftconvolve
import numpy as np
a = np.arange(30000)
b = np.ones(1000) / 1000.
%timeit fftconvolve(a, b, 'valid')
100 loops, best of 3: 6.25 ms per loop